Test for reductive hourglass patterns in transcriptomic data by comparing early and late developmental stages to mid developmental stages.
Details
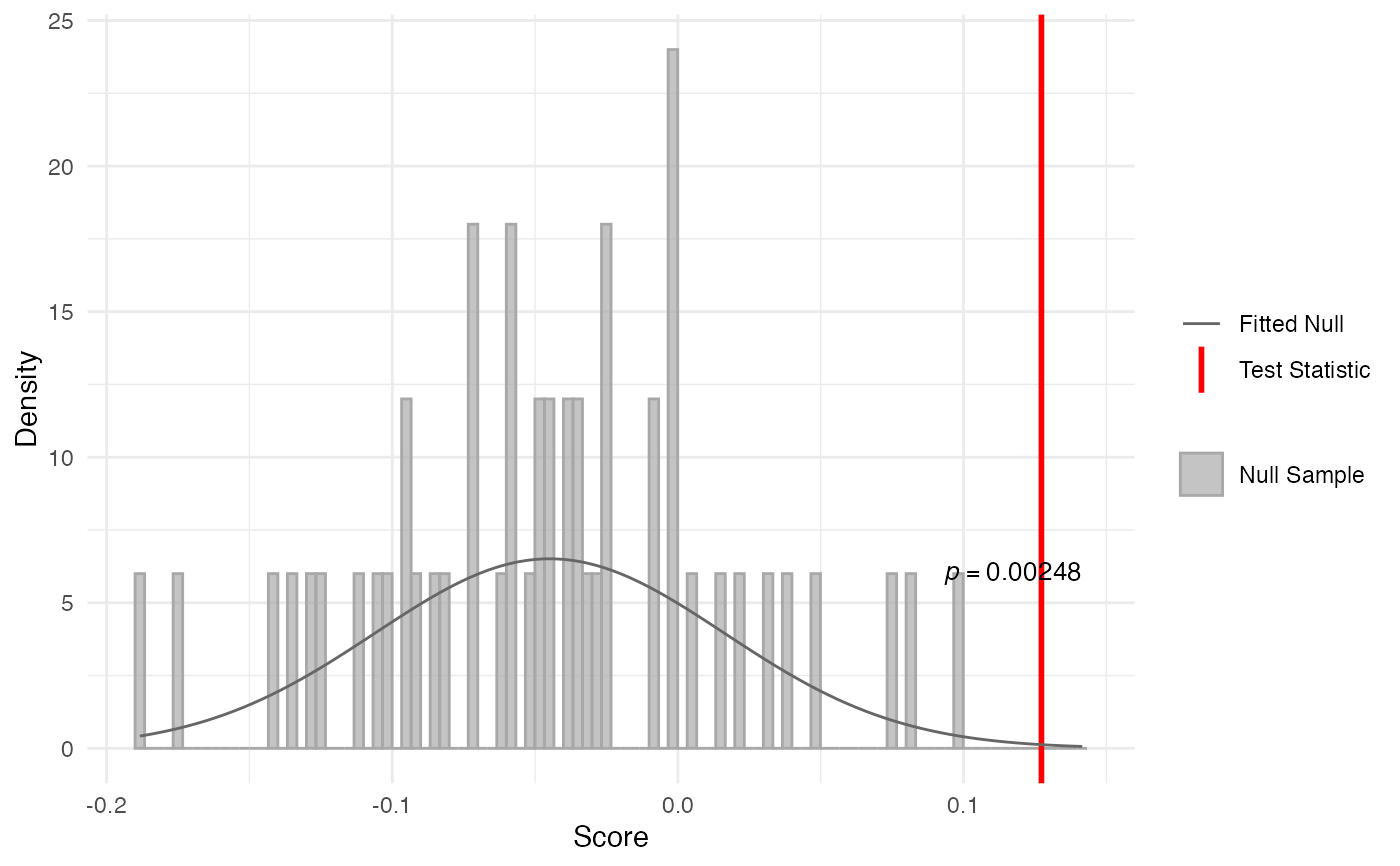
The reductive hourglass test evaluates whether mid developmental stages show lower transcriptomic index values (indicating older genes) compared to both early and late stages. This creates an hourglass-shaped pattern where ancient genes dominate during mid-development. The test computes a score based on the minimum difference between early vs. mid and late vs. mid TXI values.
Examples
# Define developmental modules
modules <- list(early = 1:2, mid = 3:5, late = 6:7)
result <- stat_reductive_hourglass_test(example_phyex_set_old, modules=modules)