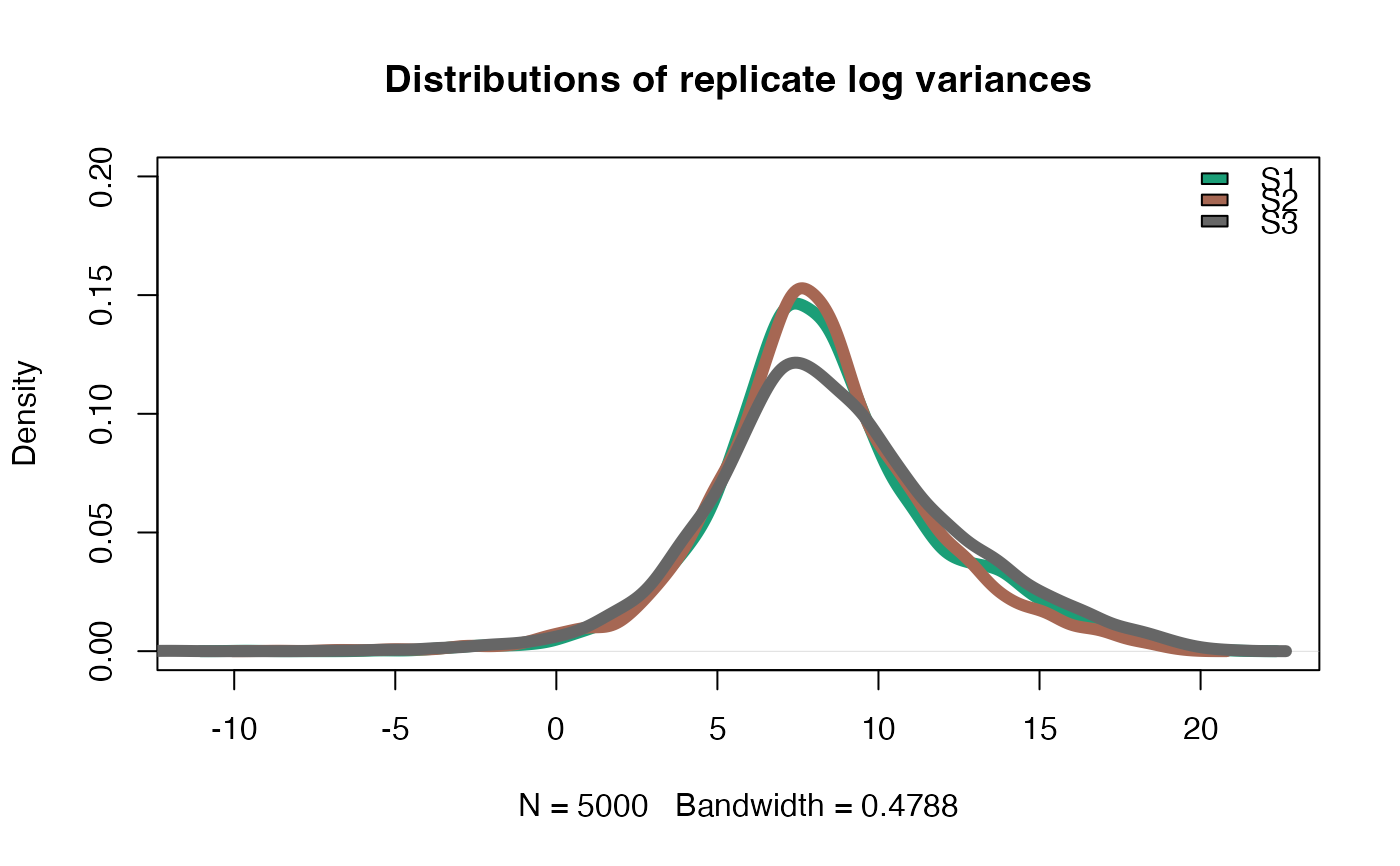
This function performs several quality checks to validate the biological variation between replicates and stages (experiments).
Arguments
- ExpressionSet
a standard PhyloExpressionSet or DivergenceExpressionSet object.
- nrep
either a numeric value specifying the constant number of replicates per stage or a numeric vector specifying the variable number of replicates for each stage position.
- FUN
a function that should be applied to quantify the variablity or quality of replicates. The default function is the log(var(x)) quantifying the variance between replicates.
- legend.pos
the position of the legend, e.g. 'topleft', or 'topright' (see
legend).- stage.names
a character vector specifying the stage names.
- ...
additional graphics parameters.