Introduction to the edgynode package
2023-11-15
Introduction.RmdTable of Contents
Why edgynode?
With the maturation of (single-cell) sequencing technologies across
modalities, the inference of gene regulatory networks (GRN)
is increasingly feasible. Yet, we lack a simple and powerful statistical
package to compare GRN inference outcomes.
The edgynode package imports inferred gene regulatory
networks and performs network statistics and network simulation
procedures to investigate the topology and structure of the GRN at
hand.
Installation
# install edgynode from GitHub
devtools::install_github("drostlab/edgynode")Performing an Example Workflow for edgynode analysis
Small example with internal dataset
# library(edgynode)
# Benchmark GENIE3 inferred networks with raw, no_noise, and quantile_norm combinations
genie3_49_raw <- as.matrix(read.csv(
system.file("data/network_raw_49_placenta_development.csv",
package = "edgynode"), row.names = 1))
genie3_49_noNoiseCM_raw <- as.matrix(read.csv(
system.file("data/network_noNoiseCM_raw_49_placenta_development.csv",
package = "edgynode"), row.names = 1))
genie3_49_qnorm_no_noise_removed <- as.matrix(read.csv(
system.file("data/network_qnorm_49_placenta_development.csv",
package = "edgynode"), row.names = 1))
genie3_49_noNoiseCM_qnorm <- as.matrix(read.csv(
system.file("data/network_noNoiseCM_qnorm_49_placenta_development.csv",
package = "edgynode"), row.names = 1))
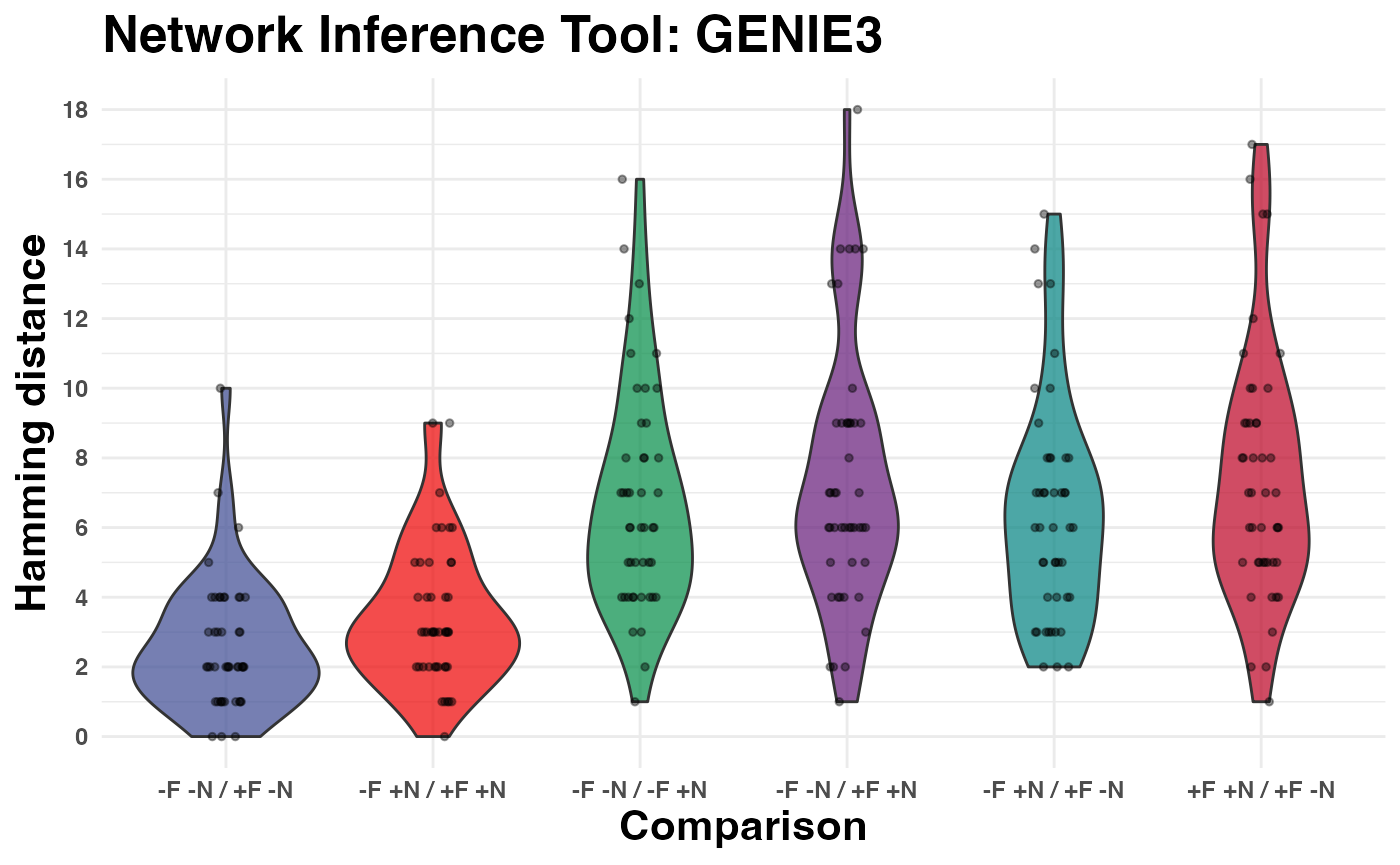
# Run Benchmark using Hamming distance
benchmark_hamming <-
edgynode::network_benchmark_noise_filtering(
genie3_49_raw,
genie3_49_noNoiseCM_raw,
genie3_49_qnorm_no_noise_removed,
genie3_49_noNoiseCM_qnorm,
dist_type = "hamming",
grn_tool = "GENIE3")
# visualize at results
edgynode::plot_network_benchmark_noise_filtering(
benchmark_hamming,
dist_type = "hamming",
title = "Network Inference Tool: GENIE3")